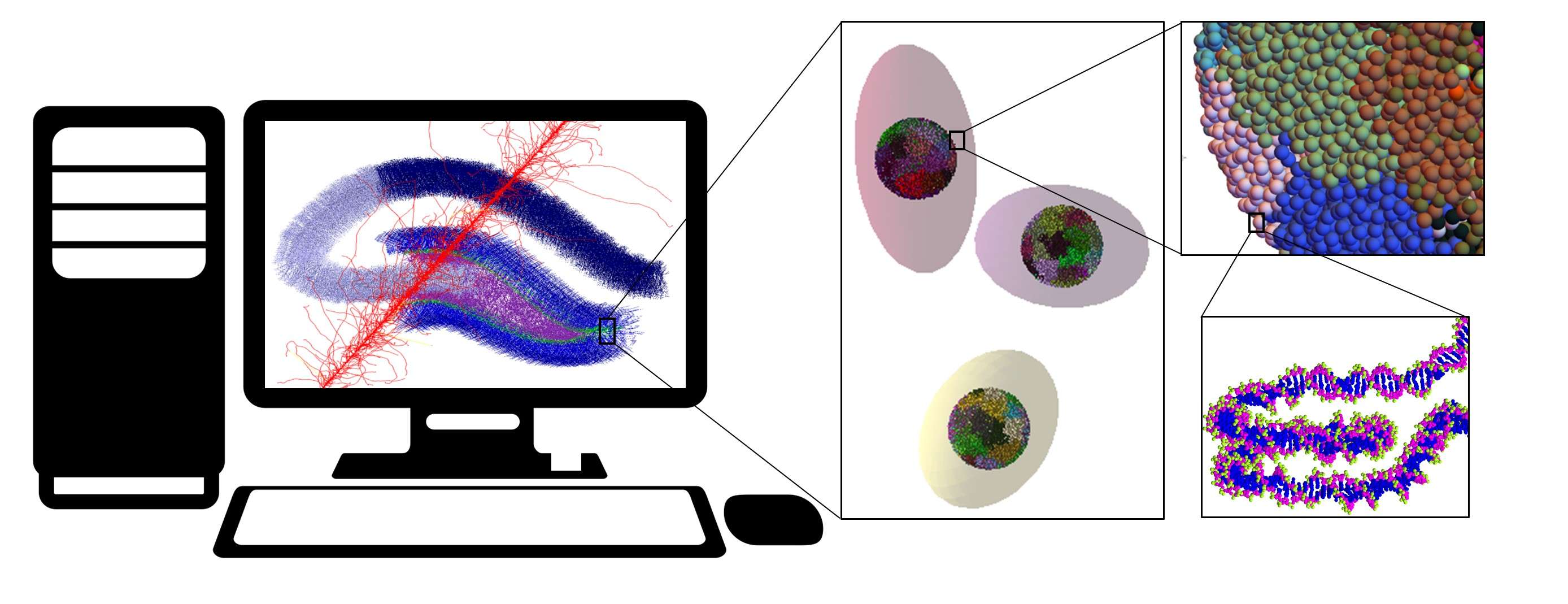
Mathematical modeling allows to describe radiation-induced effects at different levels of biological organization and time scales. Understanding these fundamental mechanisms is critical for practical applications in radiotherapy and nuclear medicine, as well as assessing health risks during interplanetary spaceflights. This project trains students in the area of radiation biophysics using Monte-Carlo codes of Geant4 and Geant4-DNA, together with specialized radiobiological models developed at the JINR Laboratory of Radiation Biology over the past 10 years.
Tasks
Individual tasks will be assigned to students based on their academic level (BSc, MSc, PhD) and prior experience. Potential tasks include:
• Simulating microscopic track structures of heavy charged particles relevant to JINR accelerators using the Geant4-DNA physics of molecular excitations, ionizations, dissociative attachments, vibrational excitation, charge exchanges, and elastic scattering.
• Building biological geometries and mapping energy depositions to DNA and subcellular targets along particle tracks.
• Investigating the influence of radiation parameters (UHDR, beam structure) on the spatial and temporal dynamics of cellular oxygen chemistry and biomolecular damage.
• Modeling radiobiological damage in different cell types and comparing outcomes.
Preliminary schedule by topics/tasks
• Literature review and software familiarization
• Hands-on training and guided simulation work
• Data analysis and scenario comparison
• Final simulation runs and visualization
• Individual report preparation
Required skills
• Basic knowledge of radiation–matter interactions and molecular radiobiology.
• Experience with Monte Carlo simulations (preferably C++ and Geant4).
• Familiarity with basic data analysis tools (ROOT) is helpful.
• Personal laptop
Acquired skills and experience
By the end of the project each student will be able to:
• Gain practical skills (hands-on experience) in particle track-structure simulation, cellular geometry construction, cluster-analysis of physico-chemical events and basic biophysical interpretation of DNA and cellular damage.
• Become familiar with Geant4 and Geant4-DNA simulation tools as well as specialized JINR radiobiological models for accelerator-based research.
• Analyze and present reproducible simulation results and prepare material suitable for a technical note or research paper.
Recommended literature
• Baeyens, A. et al. (2023). Basic Concepts of Radiation Biology. In: Baatout, S. (eds) Radiobiology Textbook. Springer, Cham. https://doi.org/10.1007/978-3-031-18810-7_2
• Batmunkh, M., Bayarchimeg, L. & Bugay, A.N. Mathematical Modeling of Radiation-Induced Effects in the Structures of the Central Nervous System under the Action of Accelerated Heavy Charged Particles. Phys. Part. Nuclei 56, 1030–1058 (2025). https://link.springer.com/article/10.1134/S1063779625700157