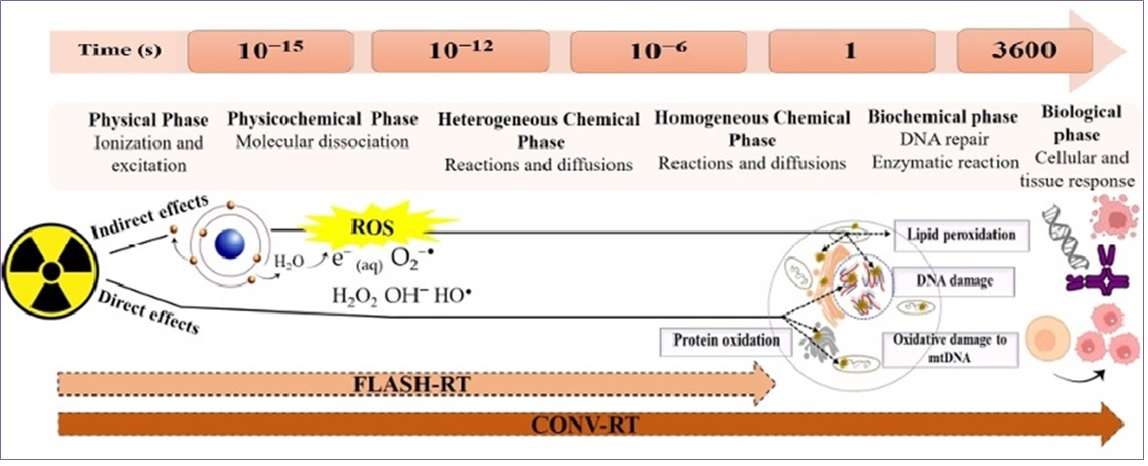
Due to the growing interest in studying the macroscopic behavior of radiation when ultra-high dose rate radiation is applied in radiotherapy (FLASH-RT), general transport codes have developed track-structured models that are used in different fields such as: radiation biology, radiotherapy, semiconductor technologies and detector physics. The track-structure calculation uses cross sections to explicitly simulate each energy deposition event attributed to atomic interaction, achieving sub-micron spatial resolution and sub-keV energy resolution. This project will focus on computer simulation of radiolytic yields for main chemical species at cellular scales and the resulting biological damage in order to compare Flash and Conventional radiotherapies for different oxygen concentrations.
Tasks
Different tasks will be assigned to BSc, MSc, PhD students, among which would be:
To reproduce experimental values of G-values (radiolytic yields) for main chemical species in water irradiated with electrons and/or gammas
To simulate the temporal kinetics of oxygen consumption during FLASH irradiation vs CONV-RT
To evaluate the differential production of reactive oxygen species under conditions of normoxia (normal tissue) and hypoxia (tumor)
To introduce the geometry of subcellular structures to evaluate the radiobiological damage at the scale from 1 micrometer to several nanometers.
Preliminary schedule by topics/tasks
Preparation and validation of the input files according to the code used
Advice and guidance in the development of computer simulations
Analysis and comparison of the results with the available experimental data
Assessment of progress and preparation of reports
Required skills
Some experience in modeling using the Monte Carlo method (preferably with the PHITS and GEANT codes)
Knowledge of basics aspects of radiation biology.
Experience with data analysis and handling of experimental databases
Acquired skills and experience
This project is aimed to acquire skills such as:
o Practical experience in Monte Carlo modeling of radiation interaction processes with subcellular structures
o Application and validation of the track-structured models to the improvement of cancer radiotherapy.
o Analysis and validation of the results of the simulations with the experimental evidences.
Recommended literature
1) Y. Matsuya et al, Modeling of yield estimation for DNA strand breaks based on Monte Carlo simulations of electron track structure in liquid water. J. of Applied Physics 126, 124701 (2019); https://doi.org/10.1063/1.5115519
2) T. Ogawa et al. “Overview of PHITS Ver.3.34 with particular focus on track-structure calculation”. EPJ Nuclear Sci. Technol. 10, 13 (2024) https://doi.org/10.1051/epjn/2024012
3) J. Schuemann et al. “TOPAS-nBio: An Extension to the TOPAS simulation Toolkit for Cellular and Sub-celullar Radiobiology”. Radiation Research 191 pag. 125 (2019). https://doi.org/10.1667/RR15226.1
4) Baeyens, A. et al. (2023). Basic Concepts of Radiation Biology. In: Baatout, S. (eds) Radiobiology Textbook. Springer, Cham.
https://doi.org/10.1007/978-3-031-18810 7_2